"Detection of SARS-CoV-2 from Wastewater"
Over the course of the COVID-19 pandemic, wastewater surveillance has become a useful tool for describing SARS-CoV-2 prevalence in populations of varying size, from individual facilities (e.g., university residence halls, nursing homes, prisons) to entire municipalities. Wastewater analysis for SARS-CoV-2 RNA requires specialized equipment, expensive consumables, and expert staff, limiting its feasibility and scalability. Further, the extremely labile nature of viral RNA complicates sample transportation, especially in regions with limited access to reliable cold chains. We work on a new method for wastewater analysis, termed exclusion-based sample preparation (ESP), that substantially simplifies workflow (at least 70% decrease in time; 40% decrease in consumable usage compared with traditional techniques) by targeting the labor-intensive processing steps of RNA purification and concentration. We have analyzed wastewater samples from residence halls at the University of Kentucky, of which 34% (44/129) contained detectible SARS-CoV-2 RNA. Although concurrent clinical testing was not comprehensive, student infections were identified in the 7 days following a positive wastewater detection in 68% of samples.
Publications: William Strike, Atena Amirsoleimani, Abisola Olaleye, Ann Noble, Kevin Lewis, Lee Faulkner, Spencer Backus, Sierra Lindeman, Katrina Eterovich, Melicity Fraley, Mohammad Dehghan Banadaki, Soroosh Torabi, Alexus Rockward, Eli Zeitlow, Matthew Liversedge, James Keck, and Scott Berry, "Development and Validation of a Simplified Method for Analysis of SARS-CoV-2 RNA in University Dormitories", ACS ES&T Water Article ASAP, https://doi.org/10.1021/acsestwater.2c00044


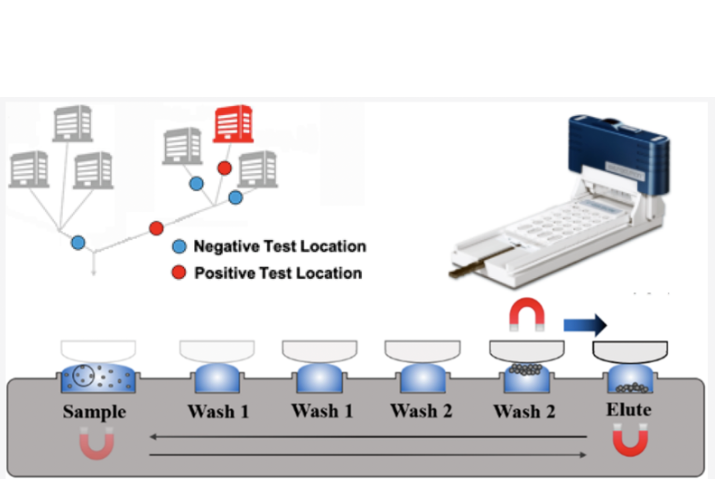
"Mobile Wastewater Assessment for Coronavirus in Kentucky"
One of our goals is to develop mobile-friendly wastewater based epidemiology methods that could be utilized in lower-resource areas. We are currently working with several locations in Appalachia, which allows us to optimize our mobile-friendly procedure and compare it to traditional, laboratory-processed results. In short, a sample is taken from the environment of interest, such as a manhole or wastewater treatment plant, and we process and purify the material into something more stable that can be measured for a disease of interest like Coronavirus.
In detail, we utilize exclusion-based sample preparation where in wastewater influent is combined with a denaturing lysis buffer such that genetic material can be bound to magnetic beads. These beads are then manipulated with magnets through wash buffers where the material is cleaned and concentrated. We then look for certain genetic sequences that are specific to COVID-19. For even more details about our protocol, please refer to our publications. In the future we hope to be able to implement public health surveillance efforts like this in even more remote and undeveloped parts of the world such as Uganda and Madagascar.
We are also beginning to develop alternate methods of wasterwater-based epidemiology that could provide further epidemic support, such as monitoring a population’s resistance to a disease of interest (via measuring for specific antibody concentrations),and tracking the mutation of a disease into different strains (via next-generation, nanopore sequencing).


"Genomic Sequencing of SARS-CoV-2 from Wastewater"
Wastewater analysis should not end at RT-qPCR analysis. Dalton have led our efforts to implement low-cost, mobile sequencing and analysis via Oxford Nanopore devices. This project was able to identify SARS-CoV-2 variants of concern in the environment at a fraction of the cost of gold-standard Next Generations Sequencing (NGS). We hope to expand our surveillance efforts from SARS-CoV-2 to drug resistant pathogens in rural Appalachia and across the globe. NGS is far too expensive and bulky to meet the needs of these populations. However, we hope to adapt Nanopore sequencing in a mobile unit for on-site library preparation and analysis.



"SARS-CoV-2 Concentration Methods"
Both symptomatic and asymptomatic patients with COVID-19 can shed the virus through the gastrointestinal tract, and this allows the detection of SARS-CoV-2 in wastewater. However, the existing viral load can become highly diluted due to the addition of more wastewater from other sources such as industrial aqueous discards, agricultural runoffs, rainfall, and other household sewage from washing, bathing, and cooking. Therefore, the concentration of the SARS-CoV-2 can fall below the limit of detection of current quantification techniques such as RT-qPCR. Also, in order to perform other quantitative methods such as ELISA and RNA sequencing, high concentrations of the target virus are needed. Therefore, we are interested in various chemical and mechanical concentration techniques to increase the number of copies per milliliter of the SARS-CoV-2 in each wastewater sample. Currently, we are developing inexpensive and easy-to-use methods to concentrate viral particles both at the point of sample collection and during the sample preparation step in the laboratory.
"SARS-CoV-2 Stability in wastewater"
One of the challenges of the wastewater-based epidemiology (WBE) of SARS-CoV-2 is the virus degradation in wastewater. The SARS-CoV-2 RNA is protected only by a lipid bilayer, which makes it susceptible to various degradation factors in wastewater such as nucleases, detergents, and heat. Therefore, it is likely to lose a large portion, if not all, of the viral load signal by the time samples are analyzed. Also, lack of reliable cold chain and lengthy wastewater sample transport times from collection points to a central lab exacerbates the degradation. Our group is investigating a SARS-CoV-2 RNA stabilization method through extraction that can be performed at the point of sample collection, which can maintain the viral RNA count for an extended period, even in the absence of refrigerated temperatures.
"Study of Rare Circulating Tumor Cells"
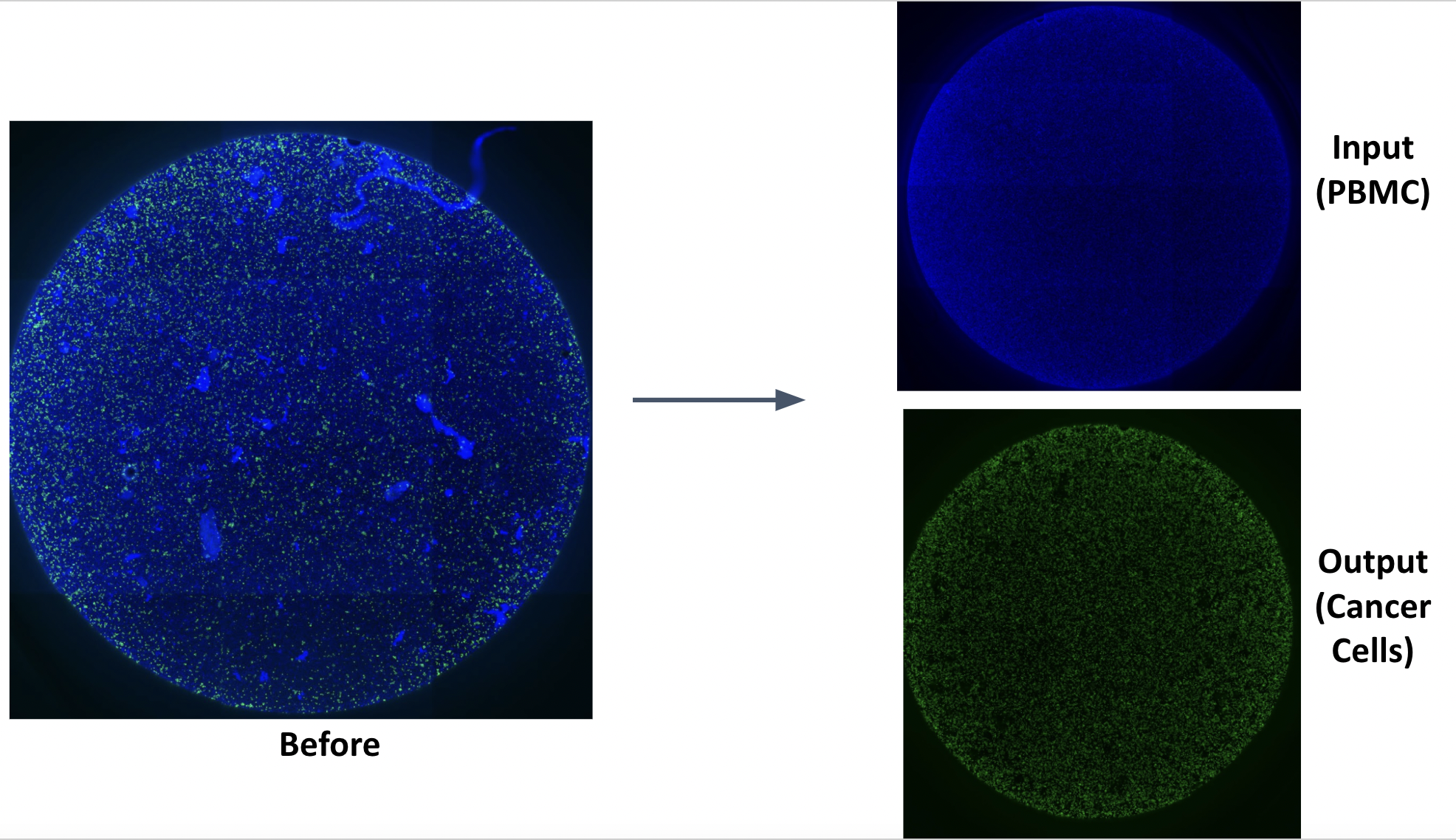
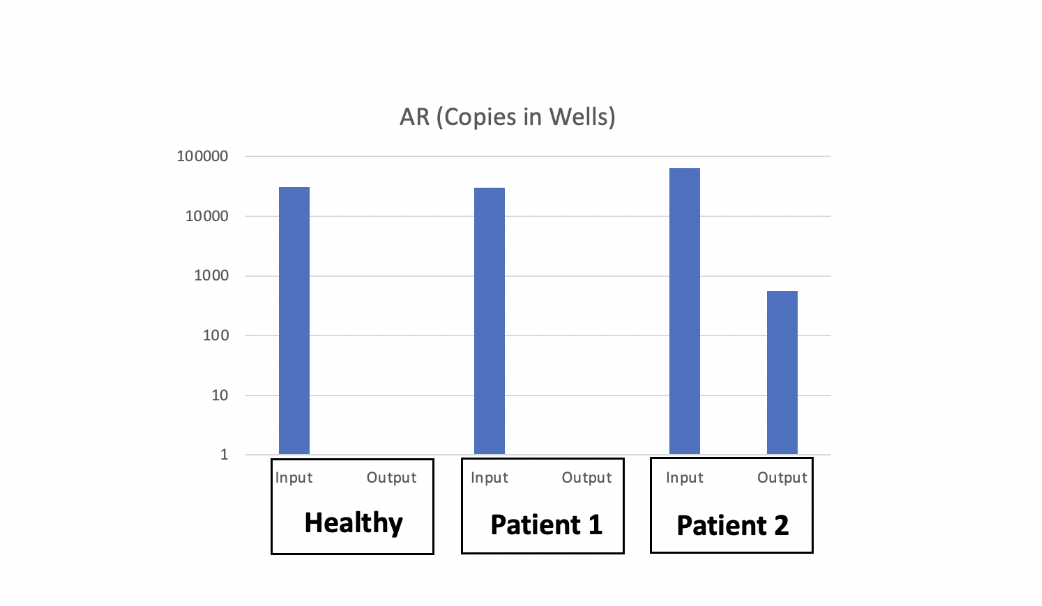
Circulating Tumor Cells (CTCs) are cancer cells that split away from the primary tumor and appear in the circulatory system. Analysis of these CTCs can capture heterogeneity across multiple sites of metastases and enables detection of changes in tumor burden that precede treatment response, and can be obtained in serial fashion. The presence of CTCs in blood offers an easy sampling method using liquid biopsy, which is far less aggressive than biopsies from tissues on the tumor site. However, these CTCs are extremely rare in the blood, making them almost invisible in the big pool of cells in the blood. We have developed a method based on exclusion-based sample preparation (ESP), that can capture and purify CTCs based on their surface molecules. In the next step, using exclusion-based sample preparation (ESP), we were able to purify the mRNA from the CTCs and study them using RT-qPCR.